Using the single template model
Quickstart example
The single template model is useful for when you know all the intrinsic parameters
of your signal (say the masses, spins, etc of a merger). In this case, we don’t
need to recalculate the waveform model to sample different possible extrinsic
parameters (i.e. distance, sky location, inclination). This can greatly
speed up the calculation of the likelihood. To use this model
you provide the intrinsic parameters as fixed arguments as in the configuration
file below.
This example demonstrates using the single_template model with the
dynesty sampler. First, we create the following configuration file:
[model]
name = single_template
#; This model precalculates the SNR time series at a fixed rate.
#; If you need a higher time resolution, this may be increased
sample_rate = 32768
low-frequency-cutoff = 30.0
[data]
instruments = H1 L1 V1
analysis-start-time = 1187008482
analysis-end-time = 1187008892
psd-estimation = median
psd-segment-length = 16
psd-segment-stride = 8
psd-inverse-length = 16
pad-data = 8
channel-name = H1:LOSC-STRAIN L1:LOSC-STRAIN V1:LOSC-STRAIN
frame-files = H1:H-H1_LOSC_CLN_4_V1-1187007040-2048.gwf L1:L-L1_LOSC_CLN_4_V1-1187007040-2048.gwf V1:V-V1_LOSC_CLN_4_V1-1187007040-2048.gwf
strain-high-pass = 15
sample-rate = 2048
[sampler]
name = dynesty
sample = rwalk
bound = multi
dlogz = 0.1
nlive = 200
checkpoint_time_interval = 100
maxcall = 10000
[variable_params]
; waveform parameters that will vary in MCMC
tc =
distance =
inclination =
[static_params]
; waveform parameters that will not change in MCMC
approximant = TaylorF2
f_lower = 30
mass1 = 1.3757
mass2 = 1.3757
#; we'll choose not to sample over these, but you could
polarization = 0
ra = 3.44615914
dec = -0.40808407
#; You could also set additional parameters if your waveform model supports / requires it.
; spin1z = 0
[prior-tc]
; coalescence time prior
name = uniform
min-tc = 1187008882.4
max-tc = 1187008882.5
[prior-distance]
#; following gives a uniform in volume
name = uniform_radius
min-distance = 10
max-distance = 60
[prior-inclination]
name = sin_angle
Download
For this example, we’ll need to download gravitational-wave data for GW170817:
set -e
for ifo in H-H1 L-L1 V-V1
do
file=${ifo}_LOSC_CLN_4_V1-1187007040-2048.gwf
test -f ${file} && continue
curl -O -L --show-error --silent https://dcc.ligo.org/public/0146/P1700349/001/${file}
done
By setting the model name to single_template we are using
SingleTemplate.
Now run:
pycbc_inference \
--config-file `dirname "$0"`/single_simple.ini \
--nprocesses=1 \
--output-file single.hdf \
--seed 0 \
--force \
--verbose
Download
This will run the dynesty sampler. When it is done, you will have a file called
single.hdf which contains the results. It should take about a minute or two to
run.
To plot the posterior distribution, run:
pycbc_inference_plot_posterior \
--input-file single.hdf \
--output-file single.png \
--z-arg snr
Download
This will create the following plot:
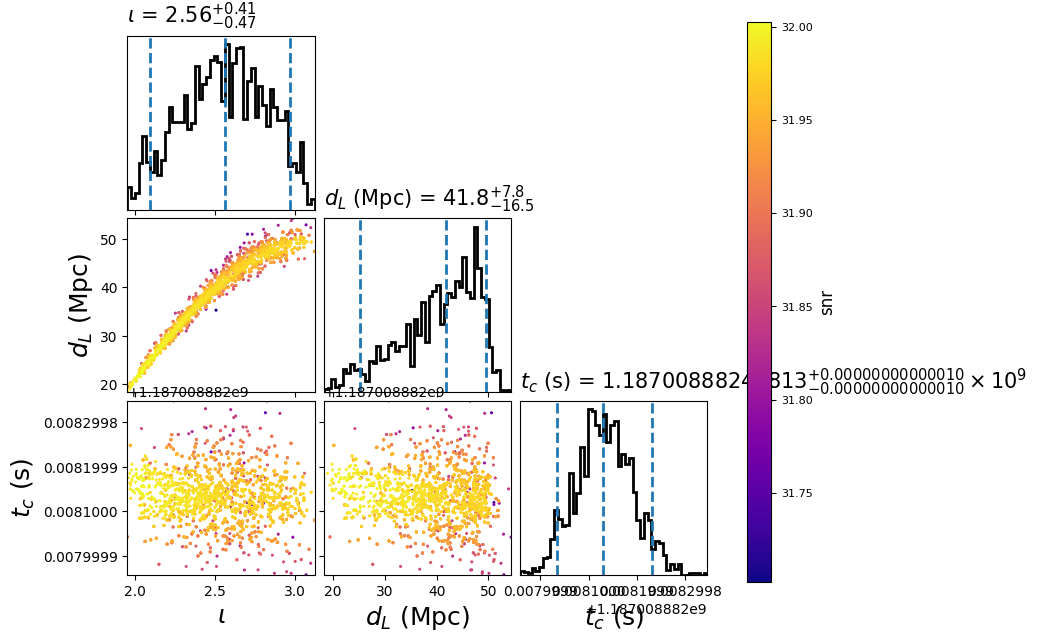
The scatter points show position of different posterior samples. The
points are colored by the log likelihood at that point, with the 50th and 90th
percentile contours drawn.
Marginalization subset of parameters
The single template model supports marginalization over its parameters. This
can greatly speed up parameter estimation. The
marginalized parameters can also be recovered as in the example below which
extends the previous example to include all the parameters the model supports.
In this example, the sampler will only explore inclination, whereas all other
parameters are either numerically or analytically marginalized over. Note
that the marginalization still takes into account the priors you choose. This
includes if you have placed boundaries or constraints.
First, you’ll need the configuration file.
[model]
name = single_template
#; This model precalculates the SNR time series at a fixed rate.
#; If you need a higher time resolution, this may be increased
sample_rate = 8192
low-frequency-cutoff = 30.0
marginalize_vector_params = tc, ra, dec, polarization
marginalize_vector_samples = 1000
# These lines enable the marginalization to only use information
# around SNR peaks for drawing possible sky / time positions. Remove
# if you want to use the entire prior range as possible
peak_lock_snr = 4.0
peak_min_snr = 4.0
marginalize_phase = True
marginalize_distance = True
marginalize_distance_param = distance
marginalize_distance_interpolator = True
marginalize_distance_snr_range = 5, 50
marginalize_distance_density = 200, 200
marginalize_distance_samples = 1000
[data]
instruments = H1 L1 V1
analysis-start-time = 1187008482
analysis-end-time = 1187008892
psd-estimation = median
psd-segment-length = 16
psd-segment-stride = 8
psd-inverse-length = 16
pad-data = 8
channel-name = H1:LOSC-STRAIN L1:LOSC-STRAIN V1:LOSC-STRAIN
frame-files = H1:H-H1_LOSC_CLN_4_V1-1187007040-2048.gwf L1:L-L1_LOSC_CLN_4_V1-1187007040-2048.gwf V1:V-V1_LOSC_CLN_4_V1-1187007040-2048.gwf
strain-high-pass = 15
sample-rate = 2048
[sampler]
name = dynesty
dlogz = 0.5
nlive = 100
[variable_params]
; waveform parameters that will vary in MCMC
#coa_phase =
distance =
polarization =
inclination =
ra =
dec =
tc =
[static_params]
; waveform parameters that will not change in MCMC
approximant = TaylorF2
f_lower = 30
mass1 = 1.3757
mass2 = 1.3757
#; we'll choose not to sample over these, but you could
#polarization = 0
#ra = 3.44615914
#dec = -0.40808407
#tc = 1187008882.42825
#; You could also set additional parameters if your waveform model supports / requires it.
; spin1z = 0
#[prior-coa_phase]
#name = uniform_angle
#[prior-ra+dec]
#name = uniform_sky
[prior-ra]
name = uniform_angle
[prior-dec]
name = cos_angle
[prior-tc]
#; coalescence time prior
name = uniform
min-tc = 1187008882.4
max-tc = 1187008882.5
[prior-distance]
#; following gives a uniform in volume
name = uniform_radius
min-distance = 10
max-distance = 60
[prior-polarization]
name = uniform_angle
[prior-inclination]
name = sin_angle
Download
Run this script to use this configuration file:
pycbc_inference \
--config-file `dirname "$0"`/single.ini \
--nprocesses=1 \
--output-file single_marg.hdf \
--seed 0 \
--force \
--verbose
pycbc_inference_plot_posterior \
--input-file single_marg.hdf \
--output-file single_marg.png \
--z-arg snr --vmin 31.85 --vmax 32.15
# This reconstructs any marginalized parameters
# and would be optional if you don't need them or
# have sampled over all parameters directly (see single.ini)
pycbc_inference_model_stats \
--input-file single_marg.hdf \
--output-file single_demarg.hdf \
--nprocesses 1 \
--reconstruct-parameters \
--force \
--verbose
pycbc_inference_plot_posterior \
--input-file single_demarg.hdf \
--output-file single_demarg.png \
--parameters distance inclination polarization coa_phase tc ra dec \
--z-arg snr --vmin 31.85 --vmax 32.15 \
Download
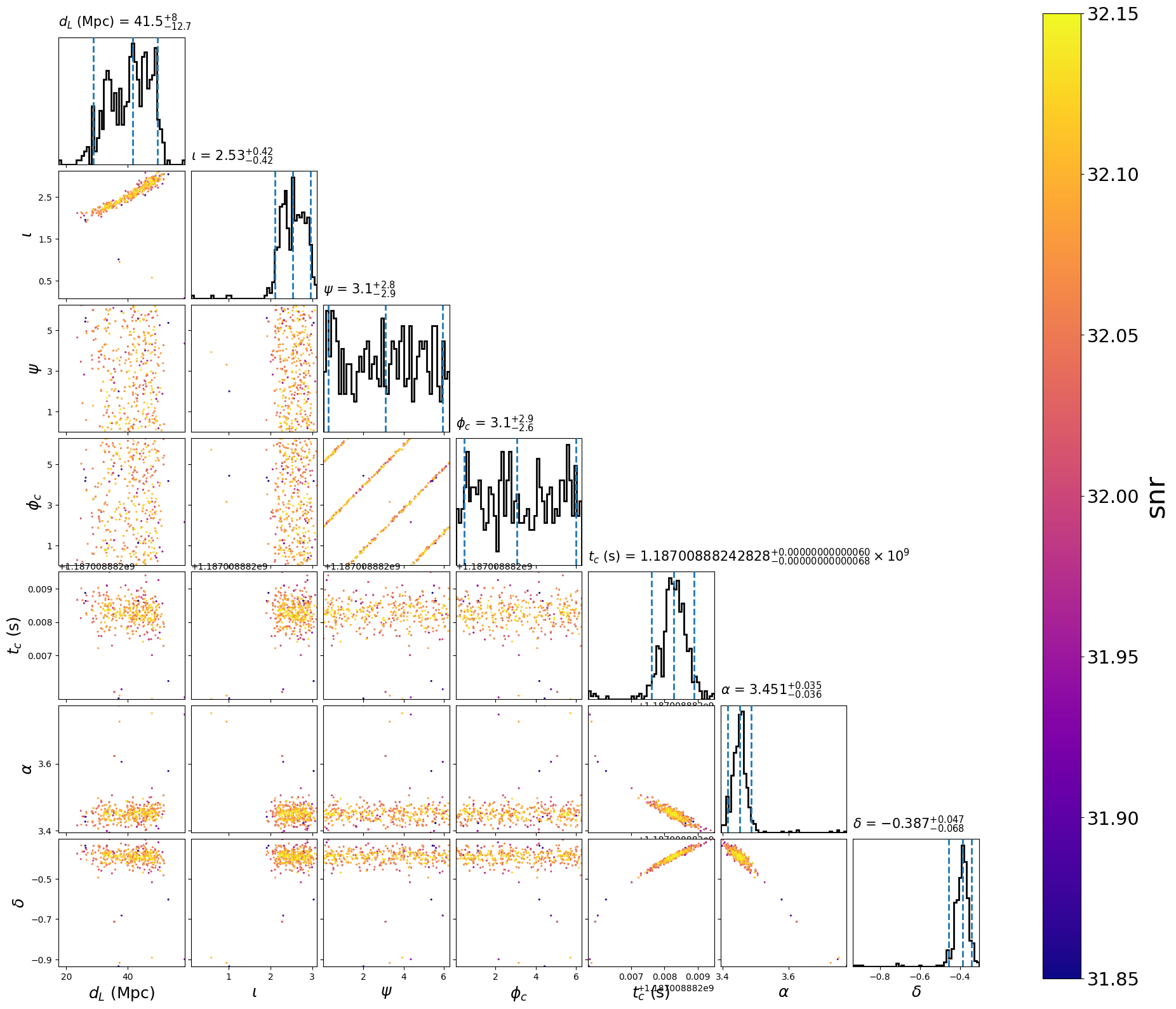
This will create the following plots:
Before demarginalization:
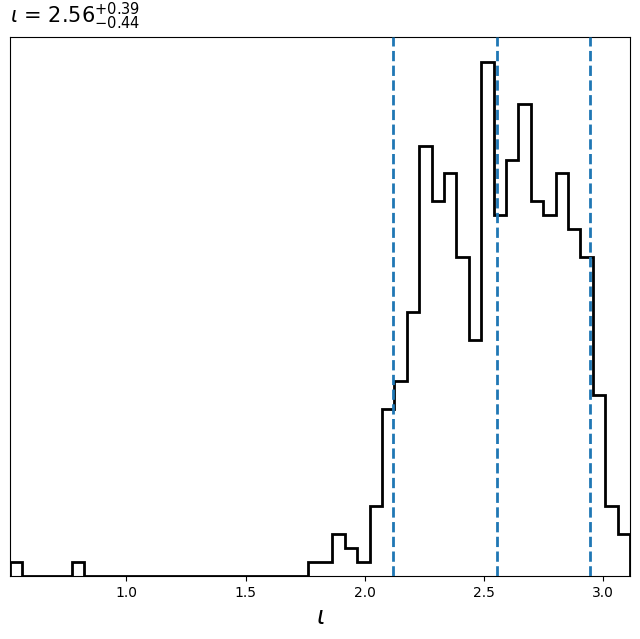
After demarginalization:
Marginalization over all parameters
All parameters that the single_template model supports can be marginalized over
to do so, you would just configure the model section to include each parameter
(minus any you choose to be static). PyCBC Inference will simply reconstruct
the missing parameters rather than performing any sampling over the parameter
space. The number of samples to take from the posterior is configurable.
First, you’ll need the configuration file. Note that in the sampler section
you can set the number of samples to draw from the posterior.
[model]
name = single_template
#; This model precalculates the SNR time series at a fixed rate.
#; If you need a higher time resolution, this may be increased
sample_rate = 16384
low-frequency-cutoff = 30.0
marginalize_vector_params = tc, ra, dec, polarization, inclination
marginalize_vector_samples = 1000
marginalize_phase = True
marginalize_distance = True
marginalize_distance_param = distance
marginalize_distance_interpolator = True
marginalize_distance_snr_range = 5, 50
marginalize_distance_density = 200, 200
marginalize_distance_samples = 1000
[data]
instruments = H1 L1 V1
analysis-start-time = 1187008482
analysis-end-time = 1187008892
psd-estimation = median
psd-segment-length = 16
psd-segment-stride = 8
psd-inverse-length = 16
pad-data = 8
channel-name = H1:LOSC-STRAIN L1:LOSC-STRAIN V1:LOSC-STRAIN
frame-files = H1:H-H1_LOSC_CLN_4_V1-1187007040-2048.gwf L1:L-L1_LOSC_CLN_4_V1-1187007040-2048.gwf V1:V-V1_LOSC_CLN_4_V1-1187007040-2048.gwf
strain-high-pass = 15
sample-rate = 2048
[sampler]
name = dummy
num_samples = 5000
[variable_params]
; waveform parameters that will vary in MCMC
#coa_phase =
distance =
polarization =
inclination =
ra =
dec =
tc =
[static_params]
; waveform parameters that will not change in MCMC
approximant = TaylorF2
f_lower = 30
mass1 = 1.3757
mass2 = 1.3757
#; we'll choose not to sample over these, but you could
#polarization = 0
#ra = 3.44615914
#dec = -0.40808407
#tc = 1187008882.42825
#; You could also set additional parameters if your waveform model supports / requires it.
; spin1z = 0
#[prior-coa_phase]
#name = uniform_angle
#[prior-ra+dec]
#name = uniform_sky
[prior-ra]
name = uniform_angle
[prior-dec]
name = cos_angle
[prior-tc]
#; coalescence time prior
name = uniform
min-tc = 1187008882.4
max-tc = 1187008882.5
[prior-distance]
#; following gives a uniform in volume
name = uniform_radius
min-distance = 10
max-distance = 60
[prior-polarization]
name = uniform_angle
[prior-inclination]
name = sin_angle
Download
Run this script to use this configuration file:
pycbc_inference \
--config-file `dirname "$0"`/single_instant.ini \
--nprocesses=1 \
--output-file single_instant.hdf \
--seed 0 \
--force \
--verbose
pycbc_inference_plot_posterior \
--input-file single_instant.hdf \
--output-file single_instant.png \
--parameters distance inclination polarization coa_phase tc ra dec \
--z-arg snr --vmin 31.85 --vmax 32.15 \
Download
This will create the following plot:
After demarginalization:
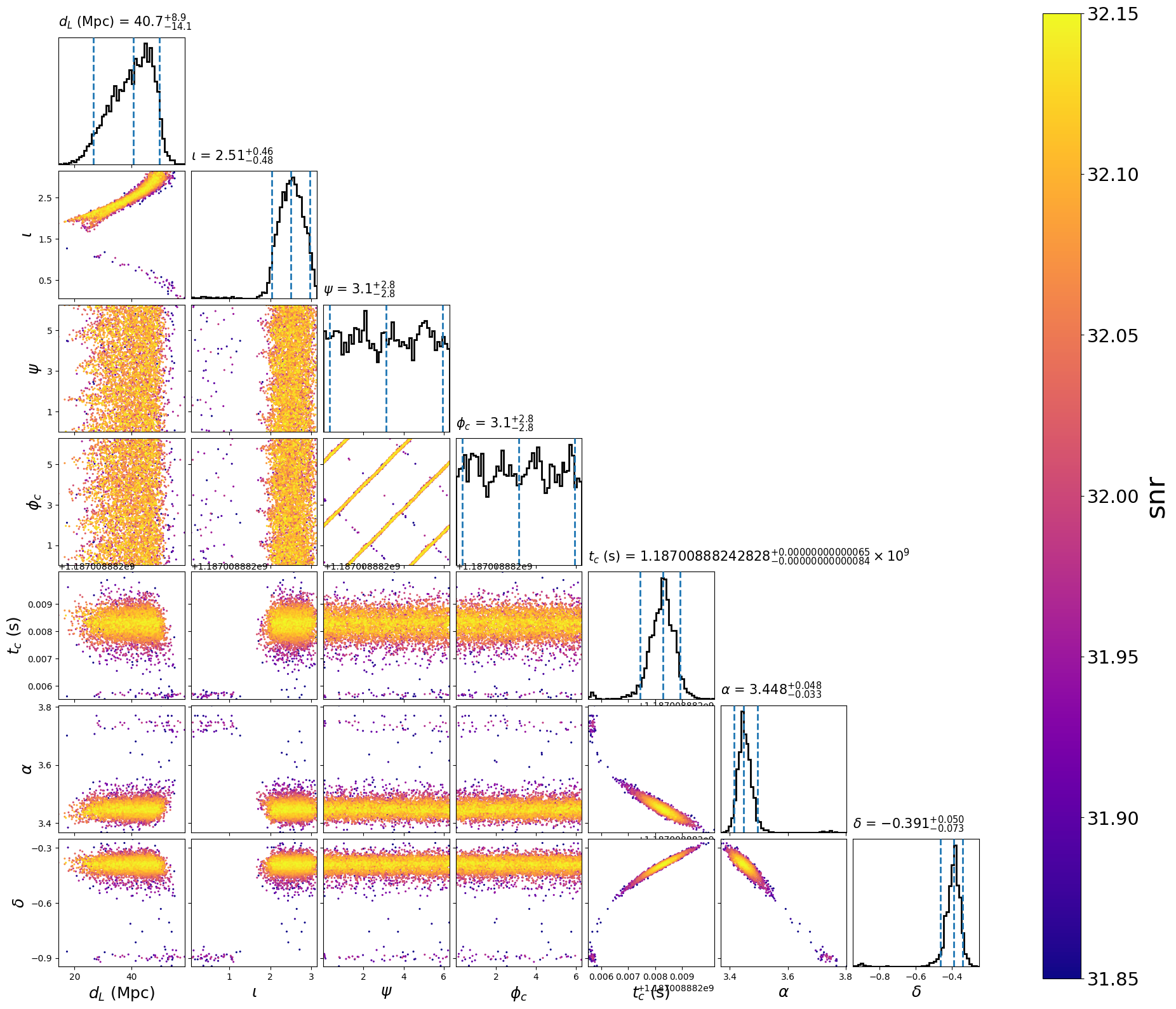
Abitrary sampling coordinates with nested samplers
The single template model also supports marginalization over the polarization
angle by numerical sampling. The following example features arbitrary sampling
coordinates with nested samplers
using the fixed_samples distribution. Here we sample in the time delay
space rather than sky location directly. This functionality is generic
to pycbc inference and could be used with other models or samplers.
[model]
name = single_template
#; This model precalculates the SNR time series at a fixed rate.
#; If you need a higher time resolution, this may be increased
sample_rate = 32768
low-frequency-cutoff = 30.0
marginalized_vector_samples = 100
marginalized_vector_params = polarization
[data]
instruments = H1 L1 V1
analysis-start-time = 1187008482
analysis-end-time = 1187008892
psd-estimation = median
psd-segment-length = 16
psd-segment-stride = 8
psd-inverse-length = 16
pad-data = 8
channel-name = H1:LOSC-STRAIN L1:LOSC-STRAIN V1:LOSC-STRAIN
frame-files = H1:H-H1_LOSC_CLN_4_V1-1187007040-2048.gwf L1:L-L1_LOSC_CLN_4_V1-1187007040-2048.gwf V1:V-V1_LOSC_CLN_4_V1-1187007040-2048.gwf
strain-high-pass = 15
sample-rate = 2048
[sampler]
name = dynesty
nlive = 100
[variable_params]
; waveform parameters that will vary in MCMC
tc =
distance =
inclination =
dh =
dhl =
[static_params]
; waveform parameters that will not change in MCMC
approximant = TaylorF2
f_lower = 30
mass1 = 1.3757
mass2 = 1.3757
#polarization = 0
[prior-tc]
; coalescence time prior
name = uniform
min-tc = 1187008882.4
max-tc = 1187008882.5
[prior-distance]
#; following gives a uniform in volume
name = uniform_radius
min-distance = 10
max-distance = 60
[prior-inclination]
name = sin_angle
[prior-dh+dhl]
name = fixed_samples
subname = mysky
sample-size = 1e6
[mysky_sample-ra+dec]
name = uniform_sky
[mysky_transform-dh+dhl]
name = custom
inputs = ra, dec
dh = det_tc('H1', ra, dec, 1187008882.0, relative=1)
dhl = det_tc('L1', ra, dec, 1187008882.0, relative=1) - det_tc('H1', ra, dec, 1187008882.0, relative=1)
[waveform_transforms-ra+dec]
name = mysky



