Inference IO
The pycbc.inference.io module provides classes and utilities for
reading and writing inference results to files. Each sampler must have a unique
class for reading/writing that sampler’s results from/to a file. These are the
sampler IO classes. Below, we provide an overview of the general structure of
these IO classes.
Overview & Guidelines
The guidelines for creating sampler IO classes are similar to the sampler
classes, though only having a single level of
inheritance is not required (since the base file class, BaseInferenceFile,
itself inherits from h5py.File). The following apply to
all sampler IO classes:
All sampler IO classes must inherit from
BaseInferenceFile. This is anabstract base classthat inherits fromh5py.File; consequently, all samplers will write to hdf files.All sampler IO classes must have a
nameattribute that is unique across all IO classes inpycbc.inference.io. This name is saved to the file and used to determine which class to read the file with when it is loaded (seeio.loadfilefor details). For example, the name attribute forEmceeFile– the IO class forEmceeEnsembleSampler– isemcee_file.Duplicate code should be avoided. If multiple samplers have common IO methods, those methods should be added to one or more support classes that the IO class can inherit from, in addition to
BaseInferenceFile. For example, all single-temperature MCMC samplers save their samples asnchains x niteraitonsdatasets. Consequently, functionality to read and write samples for single-temperature MCMC samplers is provided by theMCMCIOclass, by theMCMCIO.read_raw_samplesandMCMCIO.write_samplesmethods, respectively.The ratio of sampler IO classes to sampler classes should be 1:1. That is, each sampler should have a unique IO class and each IO class should only be used by one sampler.
The files that the sampler IO classes provide IO to are used for checkpointing by
pycbc_inference, and are the final data product that the program creates. This means that a sampler IO class must store enough information to the file such that the sampler can be initialized and continue running (either to resume for checkpoint, or to use the output of a run as the initial conditions for another run, to accumulate more points) as if the sampler had run continuously.All samples, including any statistics returned by the
model(log likelihood, log prior, etc.) should be stored as datasets (one for each parameter or statistic) to the file’ssamplesgroup. The shape of the datasets may be sampler (and run) specific.All sampler-specific data and metadata should be stored to the file’s
sampler_infogroup. What is in thesampler_infogroup and its organization may be sampler specific. This is where any additional information needed to start the sampler from a checkpoint (such as the random state) should be stored.Models may write additional information to the files. For example, data-dependent models may save the data that was analyzed to the
datagroup. See thepycbc.inference.modelsmodule for more details.
As mentioned above, the BaseInferenceFile class is the abstract base class
that defines the methods and properties that all sampler IO classes must have.
The abstract methods that IO classes must override are:
write_posterior
write_resume_point
write_sampler_metadata
Posterior Files
In addition to the sampler IO classes, there are two special classes who’s
purpose is to just store a posterior. These are the PosteriorFile and
InferenceTXTFile classes.
PosteriorFile read/writes hdf files. Like the sampler IO classes, it inherits
from BaseInferenceFile. The key difference is that all of the datasets in
the samples group are simple, flattened 1D arrays representing the
posterior. All sampler IO classes must be able to write their samples to a
PosteriorFile via their write_posterior method (which is required by
BaseInferenceFile). A PosteriorFile may or may not have other metadata in
it – sampler_info, model data, etc. – depending on the file that it was
created from. The only guaranteed property is that the samples are 1D
arrays.
Since a PosteriorFile may only contain 1D arrays of the samples, it most
likely cannot be used to resume a pycbc_inference run, as a sampler IO file
can. For example, a multi-tempered MCMC sampler would only write samples from
its coldest temperature chain to a PosteriorFile, discarding samples from the
hotter chains. A PosteriorFile is therefore not created by
pycbc_inference. Instead, pycbc_inference_extract_samples may be run
on a sampler file to produce a PosteriorFile. The primary purpose of a
PosteriorFile is to provide a compact, easy to read file that is universal
for all samplers.
The InferenceTXTFile class takes this universality one step further: it is a
simple text file containing 1D arrays of the posterior samples for each
parameter. The primary purpose of InferenceTXTFile is to provide an API for
text files that is similar to the sampler IO and PosteriorFile classes, so
that pycbc_inference_plot_posterior may be used on results from any
software suite (e.g., this is used to compare results from pycbc_inference
to posteriors published by the LIGO Scientific Collaboration).
Inheritance diagrams
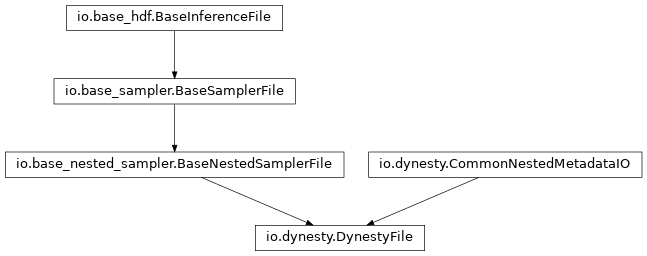
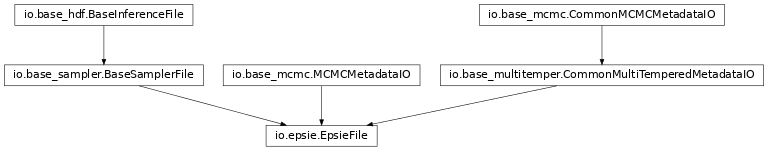
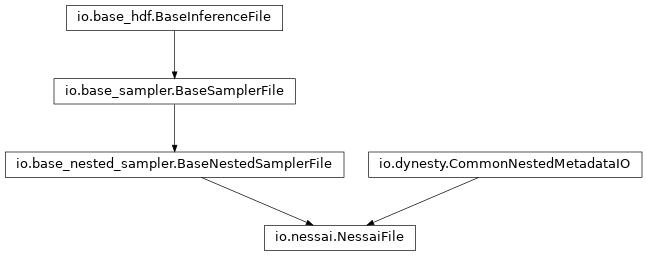
The following are inheritance diagrams for all of the currently supported IO classes:
cpnest_file:

dynesty_file:
emcee_file:

emcee_pt_file:

epsie_file:
multinest_file:

nessai_file:
posterior_file:

ptemcee_file:

snowline_file:

ultranest_file:

How to add a sampler IO class
If you are adding support for a new sampler you will also need to add a new sampler IO class. The following are steps to do that:
Create a file in
pycbc/inference/iofor the new sampler IO class.Add the new class definition. The class must inherit from at least
BaseInferenceFile.Give a name attribute to the class that is unique across the sampler IO classes.
Add any other methods you need to satisfy the
BaseInferenceFile’s required methods, plus other functionality needed to read/write relevant metadata about the sampler. You are free to write as much metadata in any format you like to thesampler_infogroup. When adding methods, try to follow the guidelines above: do not duplicate code, and try to use support classes that offer functionality that you need. If you think some of the methods will be useful for more than just your sampler, create a new support class and add those methods to it. However, if you’re unsure what is available or how best to arrange things, just add the methods you need to your new class definition. Fixing code duplication or updating support classes to can be done through the review process when you wish to add your new sampler to the main gwastro repository.Add the sampler IO class to the
filetypesdictionary inpycbc/inference/io/__init__.pyso thatio.loadfileis aware of it to use.